Informatics & Communications Centre – ICC
The ICE-HT Informatics & Communications Centre provides a wide range of IT facilities and services with a state-of-the-art networking environment to support all Institute staff in research, training, and administration.
Mission
- To facilitate the use of the latest information and communication technology in our Institute’s research, learning and administration.
- To provide an efficient and reliable network environment that facilitates the access and exchange of information.
- To put innovation in our technology transfer to assist the Institute in its mission.
- To enable our staff to use our computer and network resources effectively and efficiently.
High-Performance Computing @ FORTH/ICE-HT
Computer simulation of materials and processes constitutes one of the most active research areas at FORTH/ICE-HT. New strategies and efficient computer modeling techniques are developed and large-scale numerical calculations are performed to support and complement the experimental activities of the Institute. The modeling efforts are extended to all sectors of the Institute and provide a critical understanding of the actual mechanisms and phenomena involved in the various processes. FORTH/ICE-HT has set up a High-Performance Computing Cluster that includes a large number of powerful multicore nodes, a significant amount of memory in each node, a high-bandwidth interconnect between the nodes, optimizing compilers, and a growing set of application software.
The ICE-HT Cluster is heavily used in a variety of research fields, including air quality and climate change studies, material science and engineering, chemistry, elementary particle physics, and life sciences (e.g., systems biology, protein association and aggregation, supramolecular systems).
The Cluster has 31 compute nodes. Each compute node has two Intel Xeon two-core, quad-core, or six-core processors (5160, X5365/X5560, and E5645 respectively) and 2 GB RAM per core. That results in a total of 332 cores and 334GB of RAM available for jobs. Each compute node also provides at least 500GB of local scratch capacity on a directly attached hard disk. In addition, disk storage exceeds 20 TB, providing both high performance and archival data storage to the ICE-HT research staff. Additional login and control nodes are available for administrative purposes. The nodes are interconnected via a non-blocking Gigabit Ethernet switch providing high bandwidth and low latency for both computational communications and storage access.
Code development is implemented through Fortran C and C++ compilers from Intel and GNU Compiler Collection. The MPI libraries that are supported are Open-MPI and MPICH2. Also, the following list of scientific software packages, libraries, and other tools is installed and maintained on the Cluster:
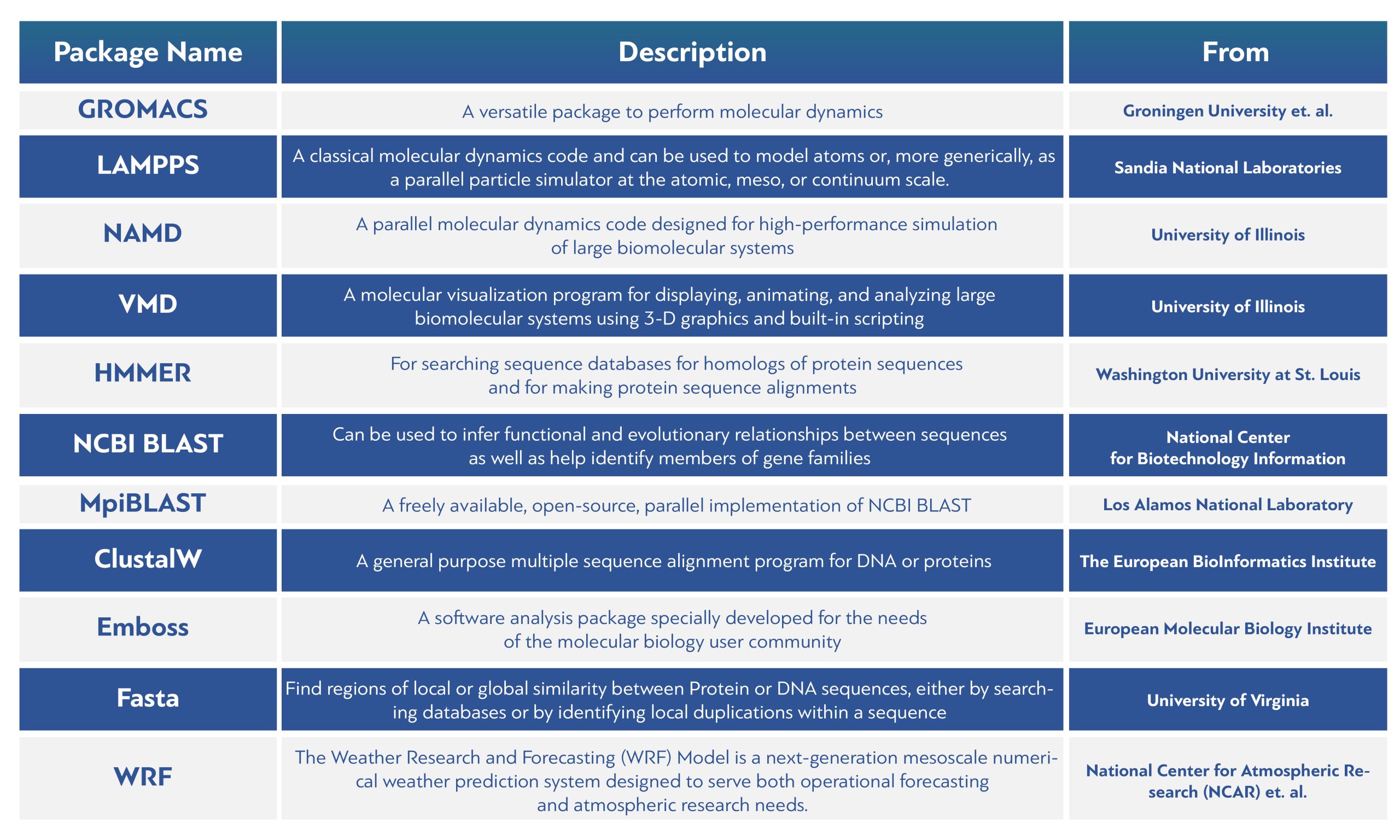
- GROMACS:A versatile package to perform molecular dynamics. From Groningen University et. al.
- LAMMPS: A classical molecular dynamics code and can be used to model atoms or, more generically, as a parallel particle simulator at the atomic, meso, or continuum scale. From Sandia National Laboratories.
- NMDA: parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. From the University of Illinois.
- VMD: A molecular visualization program for displaying, animating, and analyzing large biomolecular systems using 3-D graphics and built-in scripting. From the University of Illinois.
- HMMER: For searching sequence databases for homologs of protein sequences and for making protein sequence alignments. From the Washington University at St. Louis.
- NCBI BLAST: Can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families.From the National Center for Biotechnology Information.
- MpiBLAST: A freely available, open-source, parallel implementation of NCBI BLAST. From the Los Alamos National Laboratory.
- ClustalW: A general purpose multiple sequence alignment program for DNA or proteins. From the European BioInformatics Institute.
- Emboss:A software analysis package specially developed for the needs of the molecular biology user community.From the European Molecular Biology Institute.
- Fasta: Find regions of local or global similarity between Protein or DNA sequences, either by searching databases or by identifying local duplications within a sequence. From the University of Virginia.
- WRF: The Weather Research and Forecasting (WRF) Model is a next-generation mesoscale numerical weather prediction system designed to serve both operational forecasting and atmospheric research needs. From the National Center for Atmospheric Research (NCAR) et. al.
The ICE-HT Cluster is intended for middle-size parallel jobs that scale reasonably well for a large amount of processors and for serial jobs that have large memory requirements.
In-house Software
In addition to the aforementioned software, in-house developed software and databases are installed and run on the cluster as well as on powerful workstations. Notable examples include:
 3D Reconstruction
3D ReconstructionCode for the 3D reconstruction of porous and, in general, multiphasic materials from Electron Microscopy images
Contact: Dr. V. Burganos
 PoreFlow
PoreFlowCode for the computation of flow field in arbitrary geometries including pore structures
Contact: Dr. V. Burganos
 PoreDiffusion
PoreDiffusionCode for the computation of diffusivities and conductivities in pore structures and nanofluids
Contact: Dr. V. Burganos
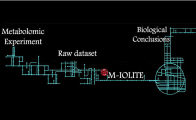 Μ-IOLITE
Μ-IOLITEIntegrated software suite for streamlining quantitative gas chromatography-mass spectrometry (GC-MS) based metabolomics analysis (supported by ELIXIR-GR)
Available for free for academic users upon request at http://miolite2.iceht.forth.gr
Contact: Dr. M.I. Klapa, Metabolic Engineering & Systems Biology Laboratory
 PICKLE: Protein InteraCtion KnowLedgebasE
PICKLE: Protein InteraCtion KnowLedgebasEMeta-database of the human and mouse experimentally-determined direct protein-protein interaction (PPI) network, integrated primary PPI databases based on genetic information ontology network (co-developed by U. of Patras and FORTH/ICE-HT; supported by ELIXIR-GR)
Available at http://www.pickle.gr
Contacts: Dr. N. K. Moschonas and Dr. M.I. Klapa
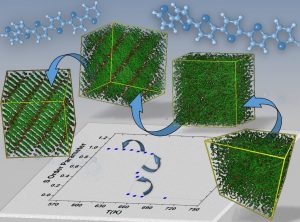 Chain connectivity altering Monte Carlo code
Chain connectivity altering Monte Carlo codeAtomistic Monte Carlo simulation code for predicting the high-temperature phase behavior of oligo- and poly-thiophenes.
Contact: Vlasis G. Mavrantzas
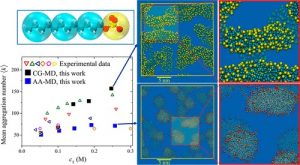 Micelles
MicellesMulti-scale particle simulation code incorporating all-atom and coarse-grained Molecular Dynamics tools for predicting micelle formation in aqueous solutions of ionic surfactants.
Contact: Vlasis G. Mavrantzas
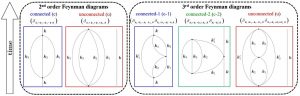 Dissipative quantum field theory
Dissipative quantum field theorySymbolic code for the calculation of multitime correlation functions in dissipative quantum field theory. Contact:
Contact: Vlasis G. Mavrantzas
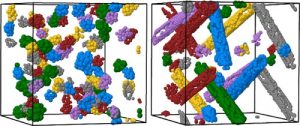 Free volume calculation
Free volume calculationGeometric code using Delaunay tessellation and Monte Carlo integration for computing the free volume and the volume accessible to small penetrants in polymer nanocomposites containing carbon nanotubes.
Contact: Vlasis G. Mavrantzas
Network Connections
The ICE-HT Campus Network is made up of a high-speed multiGigabit Ethernet resilient network backbone with over 800 network connection points on multiGigabit Ethernet segments to servers, endpoints (desktops, laptops, IP phones, tablets/smartphones), WiFi hotspots, and Virtual Private Network (VPN).
The network is connected to the Internet via EDET (Greek Academic and Research Network) that is managed by GRNET S.A., which provides high-quality services of national and international interconnection and capacity in the Greek research, academic and educational community, covering their continuously increasing requirements for high-level services and Internet applications.





